Carreté L, Ksiezopolska E, Pegueroles C, Gómez-Molero E, Saus E, Iraola S, Loska D, Bader O, Fairhead C, Gabaldón T.
“Patterns of genomic variation in the opportunistic pathogen Candida glabrata suggest the existence of mating and a secondary association with humans.”
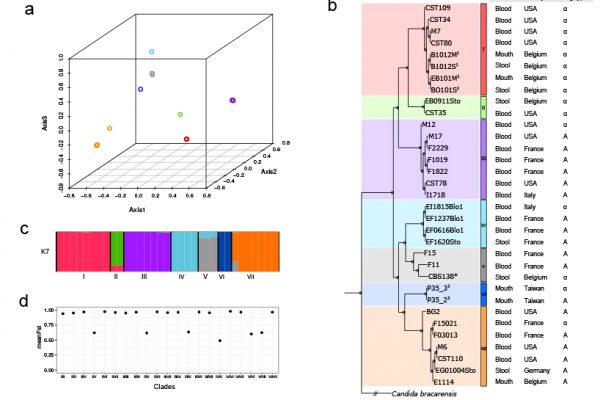
Current Biology, 28(1):15-27.e7 (2017).
Rendón-Anaya M, Montero-Vargas JM, Saburido-Álvarez S, Vlasova A, Capella-Gutierrez S, Ordaz-Ortiz JJ, Aguilar OM, Vianello-Brondani RP, Santalla M, Delaye L, Gabaldón T, Gepts P, Winkler R, Guigó R, Delgado-Salinas A, Herrera-Estrella A.
“Genomic history of the origin and domestication of common bean unveils its closest sister species.”
Genome Biol, 18(1):60 (2017).
Echeverz M, García B, Sabalza A, Valle J, Gabaldón T, Solano C, Lasa I.
“Lack of the PGA exopolysaccharide in Salmonella as an adaptive trait for survival in the host.”
PLoS Genet, 13(5):e1006816 (2017).
Masgrau A, Battola A, Sanmartin T, Pryszcz LP, Gabaldón T, Mendoza M.
“Distinct roles of the polarity factors Boi1 and Boi2 in the control of exocytosis and abscission in budding yeast.”
Mol Biol Cell, 28(22):3082-3094 (2017).
Mixão V, Gabaldón T.
“Hybridization and emergence of virulence in opportunistic human yeast pathogens.”
Yeast. 35(1):5-20 (2017).