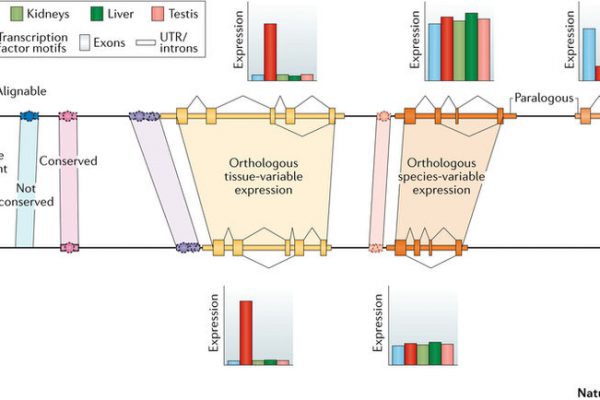
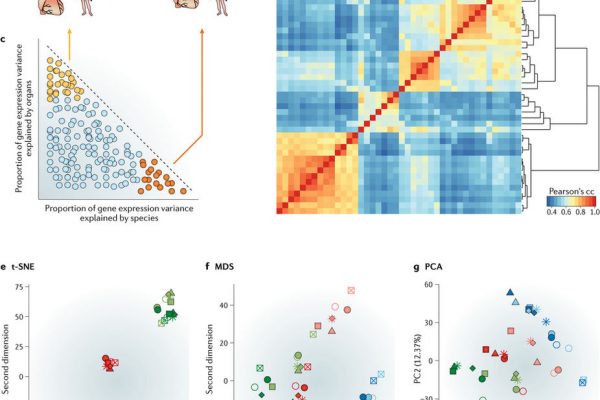
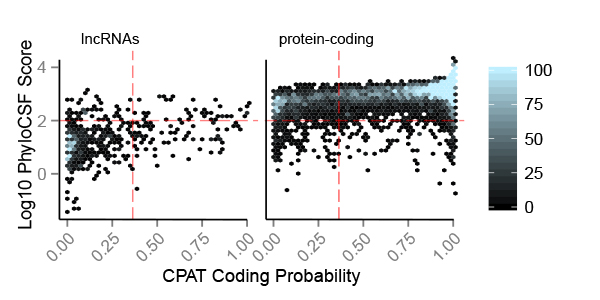
GROUP LEADER: Roderic Guigó (Coordinator of the Bionformatics and Genomics Programme)
STAFF SCIENTIST: Silvia Pérez-Lluch
POSTDOCTORAL FELLOWS: Ferran Reverter, Cecilia Klein, Marina Ruiz, Ramil Nurtdinov, Bruna Correa, Valentin Wucher, Estel Aparicio (until March), Didac Santesmasses (until June), Gireesh Kumar (until July), Alessandra Breschi (until August)
PHD STUDENTS: Sebastian Ullrich, Reza Sodaei, Diego Garrido, Andrés Arturo Lanzos, Manuel Muñoz, Beatrice Borsari
BIOINFORMATICIANS: Julien Lagarde, Francisco Câmara, Emilio Palumbo, Anna Vlasova, Joao Curado
TECHNICIANS: Carmen Arnán, Amaya Abad
STUDENTS: Raziel Tomás, Aida Ripoll, David Mas
VISITING SCIENTISTS: Lourdes Pena, Denis Tagu