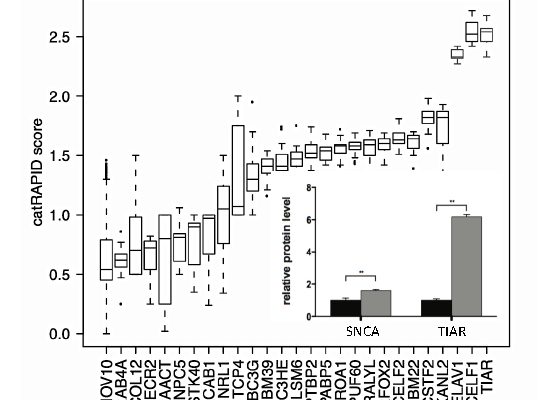
Marchese M, Botta-Orfila T, Cirillo D, Rodriguez JA, Livi CA, Fernández-Santiago R, Ezquerra M, Martí MJ Bechara E and Tartaglia GG.
“Discovering the 3’ UTR-Mediated Regulation of Alpha-Synuclein.”
Nucleic Acids Research, 10.1093/nar/gkx1048 (in press) (2017).
Ribeiro DM, Zanzoni A, Cipriano A, Ballarino M, Spinelli L, Delli Ponti R, Bozzoni I, Tartaglia GG (corresponding author), Brun C.
“Protein complex scaffolding as a prevalent function of human long non-coding RNAs.”
Nucleic Acids Research, doi: 10.1093/nar/gkx1169 (in press) (2018).
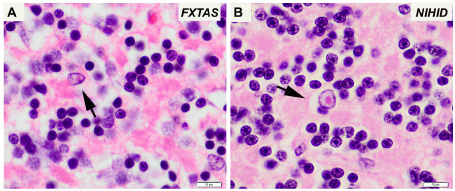
Gelpi E, Botta-Orfila T, Bodi L, Marti S, Kovacs G, Grau-Rivera O, Lozano M, Sánchez-Valle R, Muñoz E, Valldeoriola F, Pagonabarraga J, Tartaglia GG, Milà M.
“Neuronal intranuclear (hyaline) inclusion disease and fragile X-associated tremor/ataxia syndrome: a morphological and molecular dilemma.”
Brain, 140(8):e51. doi: 10.1093/brain/awx156 (2017).
Armaos A, Cirillo D, Tartaglia GG.
“omiXcore: a web server for prediction of protein interactions with large RNA.”
Bioinformatics, 33(19):3104-3106. doi: 10.1093/bioinformatics/btx361 (2017)
Cirillo D, Blanco M, Armaos A, Buness A, Avner P, Guttman M, Cerase A and Tartaglia GG.
“Quantitiative predicitons of protein interactions with long non-coding RNAs.”
Nat Methods, 14(1):5-6. doi: 10.1038/nmeth.4100 (2017).