How do networks of genes control populations of cells to build organized tissues and organs? Organ development is a multi-scale process. At the molecular scale, regulation of genes and proteins creates complex networks which control the activities of cells (cell division, migration, cell fate decisions, differentiation, and many other activities). This molecular control system also includes an extracellular part – secreted ligands which move between cells allowing cell-cell communication (such as FGFs, WNTs, etc.). The coordination of thousands of cells by this extended molecular network, leads to large-scale morphogenesis at the scale of tissues and organs. However, these “macroscopic” movements also feedback to the molecular scale: the movement of tissue regions relative to each other causes cells to receive dynamically changing concentrations of signalling molecules, and this in turn changes the activation or repression of genes.
A full understanding of how genes control organogenesis will require multi-scale computer modelling, and we have chosen vertebrate limb development as a model system to explore this problem. Such a model should be based on quantitative data about dynamic tissue shapes and spatial distributions of gene activities. Traditional high-throughput and “omics” technologies do not preserve spatial information, and we have therefore been developing and pioneering various 3D mesoscopic imaging technologies to generate geometric and spatial data for the model. During 2015, the lab was awarded an ERC Advanced Grant for continuing this work.
Our research thus falls primarily into two main areas:
(1) To further our understanding of organogenesis as a complex system, by bringing together a diverse range of techniques from biology, physics, imaging and computer science. Within this general theme, we focus on two aspects:
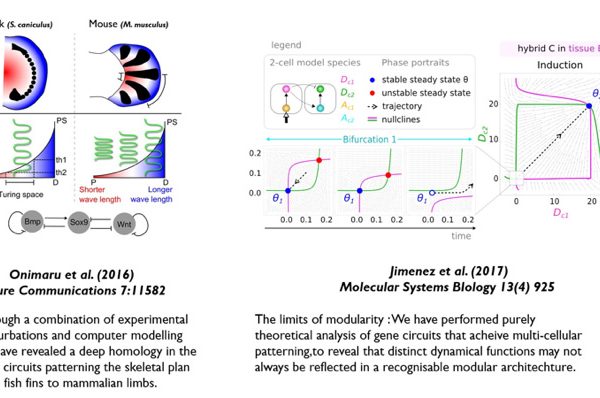
(a) We study a well-characterised standard model of development – the vertebrate limb/fin (using the mouse, chick and catshark). We combine experimental data (especially 3D data sets using optical projection tomography) within computational frameworks, to explore and test mechanistic hypotheses about how limb morphogenesis works. Using this approach, we are studying both the physical morphogenesis (eg. Boehm et al, PLoS Biol., 2010, and Marcon et al, PLoS Comp. Biol. 2011) and also the genetic patterning mechanisms (eg. Sheth et al, Science 2012, and Uzkudun et al. Molecular Systems Biology 2015). We have used this mix of experimental and theoretical work to find evidence supporting the idea that digit patterning is achieved by a Turing reaction-diffusion system (Raspopovic et al, Science 2014), and most recently have shown that this particular molecular system has been conserved all the way from fish to mammals (Onimaru et al, Nature Communications 2016).
(b) In addition to this specific model system, we are also interested in the theoretical principles by which gene regulatory networks can create controlled spatial patterns in multicellular contexts (eg. Cotterell et al, Molecular Systems Biology 2010, Schaerli et al. Nature Communications 2014), and during 2015 we have published two further studies on this theme: Cotterell et al. Cell Systems 2015, and Jimenez et al. PNAS 2015
(2) Building on the success of the 3D imaging technique developed within the lab called Optical Projection Tomography (OPT – Science 296:541, 2002), the other major goal of the lab is to continue developing and improving 3D and 4D imaging technology including the development of time-lapse imaging of mouse limb development in-vitro (eg. Nature Methods 5:609-12, 2008). Our recent projects include the development of OPTiSPIM (Maeyer et al., 2014).